import numpy as np
import utils
import json
import pandas as pd
import utils
from scipy.stats import kendalltau
import sys
import os
import math 8 Estimate \(S\) with Soft K-Means
The basic idea of using Soft K-Means to estimate \(S\) is:
- We first estimate distribution parameters using hard K-Means, the exact procedure we covered in Section 4.1.
- In each iteration, we use Soft Kmeans algorithm to update \(\theta, \phi\) (refer to Section 4.3) and use Metropolis–Hastings algorithm to accept or reject a proposed \(S\).
8.1 Implementation
Code
def calculate_soft_kmeans_for_biomarker(
data,
biomarker,
order_dict,
n_participants,
non_diseased_participants,
hashmap_of_normalized_stage_likelihood_dicts,
diseased_stages,
seed=None
):
"""
Calculate mean and std for both the affected and non-affected clusters for a single biomarker.
Parameters:
data (pd.DataFrame): The data containing measurements.
biomarker (str): The biomarker to process.
order_dict (dict): Dictionary mapping biomarkers to their order.
n_participants (int): Number of participants in the study.
non_diseased_participants (list): List of non-diseased participants.
hashmap_of_normalized_stage_likelihood_dicts (dict): Hash map of
dictionaries containing stage likelihoods for each participant.
diseased_stages (list): List of diseased stages.
seed (int, optional): Random seed for reproducibility.
Returns:
tuple: Means and standard deviations for affected and non-affected clusters.
"""
if seed is not None:
# Set the seed for numpy's random number generator
rng = np.random.default_rng(seed)
else:
rng = np.random
# DataFrame for this biomarker
biomarker_df = data[
data['biomarker'] == biomarker].reset_index(
drop=True).sort_values(
by = 'participant', ascending = True)
# Extract measurements
measurements = np.array(biomarker_df['measurement'])
this_biomarker_order = order_dict[biomarker]
affected_cluster = []
non_affected_cluster = []
for p in range(n_participants):
if p in non_diseased_participants:
non_affected_cluster.append(measurements[p])
else:
if this_biomarker_order == 1:
affected_cluster.append(measurements[p])
else:
normalized_stage_likelihood_dict = hashmap_of_normalized_stage_likelihood_dicts[
p]
# Calculate probabilities for affected and non-affected states
affected_prob = sum(
normalized_stage_likelihood_dict[s] for s in diseased_stages if s >= this_biomarker_order
)
non_affected_prob = sum(
normalized_stage_likelihood_dict[s] for s in diseased_stages if s < this_biomarker_order
)
if affected_prob > non_affected_prob:
affected_cluster.append(measurements[p])
elif affected_prob < non_affected_prob:
non_affected_cluster.append(measurements[p])
else:
# Assign to either cluster randomly if probabilities are equal
if rng.random() > 0.5:
affected_cluster.append(measurements[p])
else:
non_affected_cluster.append(measurements[p])
# Compute means and standard deviations
theta_mean = np.mean(affected_cluster) if affected_cluster else np.nan
theta_std = np.std(affected_cluster) if affected_cluster else np.nan
phi_mean = np.mean(
non_affected_cluster) if non_affected_cluster else np.nan
phi_std = np.std(non_affected_cluster) if non_affected_cluster else np.nan
return theta_mean, theta_std, phi_mean, phi_std
def soft_kmeans_theta_phi_estimates(
iteration,
prior_theta_phi_estimates,
data_we_have,
biomarkers,
order_dict,
n_participants,
non_diseased_participants,
hashmap_of_normalized_stage_likelihood_dicts,
diseased_stages,
seed=None):
"""
Get the DataFrame of theta and phi using the soft K-means algorithm for all biomarkers.
Parameters:
data_we_have (pd.DataFrame): DataFrame containing the data.
biomarkers (list): List of biomarkers in string.
order_dict (dict): Dictionary mapping biomarkers to their order.
n_participants (int): Number of participants in the study.
non_diseased_participants (list): List of non-diseased participants.
hashmap_of_normalized_stage_likelihood_dicts (dict): Hash map of dictionaries containing stage likelihoods for each participant.
diseased_stages (list): List of diseased stages.
seed (int, optional): Random seed for reproducibility.
Returns:
a dictionary containing the means and standard deviations for theta and phi for each biomarker.
"""
# List of dicts to store the estimates
# In each dic, key is biomarker, and values are theta and phi params
hashmap_of_means_stds_estimate_dicts = {}
for biomarker in biomarkers:
dic = {'biomarker': biomarker}
prior_theta_phi_estimates_biomarker = prior_theta_phi_estimates[biomarker]
theta_mean, theta_std, phi_mean, phi_std = calculate_soft_kmeans_for_biomarker(
data_we_have,
biomarker,
order_dict,
n_participants,
non_diseased_participants,
hashmap_of_normalized_stage_likelihood_dicts,
diseased_stages,
seed
)
if theta_std == 0 or math.isnan(theta_std):
theta_mean = prior_theta_phi_estimates_biomarker['theta_mean']
theta_std = prior_theta_phi_estimates_biomarker['theta_std']
if phi_std == 0 or math.isnan(phi_std):
phi_mean = prior_theta_phi_estimates_biomarker['phi_mean']
phi_std = prior_theta_phi_estimates_biomarker['phi_std']
dic['theta_mean'] = theta_mean
dic['theta_std'] = theta_std
dic['phi_mean'] = phi_mean
dic['phi_std'] = phi_std
hashmap_of_means_stds_estimate_dicts[biomarker] = dic
return hashmap_of_means_stds_estimate_dicts
def calculate_all_participant_ln_likelihood_and_update_hashmap(
iteration,
data_we_have,
current_order_dict,
n_participants,
non_diseased_participant_ids,
theta_phi_estimates,
diseased_stages,
):
data = data_we_have.copy()
data['S_n'] = data.apply(
lambda row: current_order_dict[row['biomarker']], axis=1)
all_participant_ln_likelihood = 0
# key is participant id
# value is normalized_stage_likelihood_dict
hashmap_of_normalized_stage_likelihood_dicts = {}
for p in range(n_participants):
pdata = data[data.participant == p].reset_index(drop=True)
if p in non_diseased_participant_ids:
this_participant_likelihood = utils.compute_likelihood(
pdata, k_j=0, theta_phi=theta_phi_estimates)
this_participant_ln_likelihood = np.log(
this_participant_likelihood + 1e-10)
else:
# normalized_stage_likelihood_dict = None
# initiaze stage_likelihood
stage_likelihood_dict = {}
for k_j in diseased_stages:
kj_likelihood = utils.compute_likelihood(
pdata, k_j, theta_phi_estimates)
# update each stage likelihood for this participant
stage_likelihood_dict[k_j] = kj_likelihood
# Add a small epsilon to avoid division by zero
likelihood_sum = sum(stage_likelihood_dict.values())
epsilon = 1e-10
if likelihood_sum == 0:
# print("Invalid likelihood_sum: zero encountered.")
likelihood_sum = epsilon # Handle the case accordingly
normalized_stage_likelihood = [
l/likelihood_sum for l in stage_likelihood_dict.values()]
normalized_stage_likelihood_dict = dict(
zip(diseased_stages, normalized_stage_likelihood))
hashmap_of_normalized_stage_likelihood_dicts[p] = normalized_stage_likelihood_dict
# calculate weighted average
this_participant_likelihood = np.mean(likelihood_sum)
this_participant_ln_likelihood = np.log(
this_participant_likelihood)
all_participant_ln_likelihood += this_participant_ln_likelihood
return all_participant_ln_likelihood, hashmap_of_normalized_stage_likelihood_dicts
def metropolis_hastings_soft_kmeans(
data_we_have,
iterations,
n_shuffle,
):
'''Implement the metropolis-hastings algorithm using soft kmeans
Inputs:
- data: data_we_have
- iterations: number of iterations
- log_folder_name: the folder where log files locate
Outputs:
- best_order: a numpy array
- best_likelihood: a scalar
'''
n_participants = len(data_we_have.participant.unique())
biomarkers = data_we_have.biomarker.unique()
n_biomarkers = len(biomarkers)
n_stages = n_biomarkers + 1
non_diseased_participant_ids = data_we_have.loc[
data_we_have.diseased == False].participant.unique()
diseased_stages = np.arange(start=1, stop=n_stages, step=1)
# obtain the iniial theta and phi estimates
prior_theta_phi_estimates = utils.get_theta_phi_estimates(
data_we_have)
theta_phi_estimates = prior_theta_phi_estimates.copy()
# initialize empty lists
acceptance_count = 0
all_current_accepted_order_dicts = []
current_accepted_order = np.random.permutation(np.arange(1, n_stages))
current_accepted_order_dict = dict(zip(biomarkers, current_accepted_order))
current_accepted_likelihood = -np.inf
for _ in range(iterations):
# in each iteration, we have updated current_order_dict and theta_phi_estimates
new_order = current_accepted_order.copy()
utils.shuffle_order(new_order, n_shuffle)
current_order_dict = dict(zip(biomarkers, new_order))
all_participant_ln_likelihood, \
hashmap_of_normalized_stage_likelihood_dicts = calculate_all_participant_ln_likelihood_and_update_hashmap(
_,
data_we_have,
current_order_dict,
n_participants,
non_diseased_participant_ids,
theta_phi_estimates,
diseased_stages,
)
# Now, update theta_phi_estimates using soft kmeans
# based on the updated hashmap of normalized stage likelihood dicts
theta_phi_estimates = soft_kmeans_theta_phi_estimates(
_,
prior_theta_phi_estimates,
data_we_have,
biomarkers,
current_order_dict,
n_participants,
non_diseased_participant_ids,
hashmap_of_normalized_stage_likelihood_dicts,
diseased_stages,
seed=None,
)
# Log-Sum-Exp Trick
max_likelihood = max(all_participant_ln_likelihood,
current_accepted_likelihood)
prob_of_accepting_new_order = np.exp(
(all_participant_ln_likelihood - max_likelihood) -
(current_accepted_likelihood - max_likelihood)
)
# prob_of_accepting_new_order = np.exp(
# all_participant_ln_likelihood - current_accepted_likelihood)
# np.exp(a)/np.exp(b) = np.exp(a - b)
# if a > b, then np.exp(a - b) > 1
# it will definitly update at the first iteration
if np.random.rand() < prob_of_accepting_new_order:
acceptance_count += 1
current_accepted_order = new_order
current_accepted_likelihood = all_participant_ln_likelihood
current_accepted_order_dict = current_order_dict
acceptance_ratio = acceptance_count*100/(_+1)
all_current_accepted_order_dicts.append(current_accepted_order_dict)
if (_+1) % 10 == 0:
formatted_string = (
f"iteration {_ + 1} done, "
f"current accepted likelihood: {current_accepted_likelihood}, "
f"current acceptance ratio is {acceptance_ratio:.2f} %, "
f"current accepted order is {current_accepted_order_dict.values()}, "
)
print(formatted_string)
# print("done!")
return all_current_accepted_order_dictsn_shuffle = 2
iterations = 10
burn_in = 2
thining = 2
base_dir = os.getcwd()
print(f"Current working directory: {base_dir}")
data_dir = os.path.join(base_dir, "data")
soft_kmeans_dir = os.path.join(base_dir, 'soft_kmeans')
temp_results_dir = os.path.join(soft_kmeans_dir, "temp_json_results")
img_dir = os.path.join(soft_kmeans_dir, 'img')
results_file = os.path.join(soft_kmeans_dir, "results.json")
os.makedirs(soft_kmeans_dir, exist_ok=True)
os.makedirs(temp_results_dir, exist_ok=True)
os.makedirs(img_dir, exist_ok=True)
print(f"Data directory: {data_dir}")
print(f"Temp results directory: {temp_results_dir}")
print(f"Image directory: {img_dir}")
if __name__ == "__main__":
# Read parameters from command line arguments
j = 200
r = 0.75
m = 3
print(f"Processing with j={j}, r={r}, m={m}")
combstr = f"{int(j*r)}|{j}"
heatmap_folder = img_dir
img_filename = f"{int(j*r)}-{j}_{m}"
filename = f"{combstr}_{m}"
data_file = f"{data_dir}/{filename}.csv"
data_we_have = pd.read_csv(data_file)
n_biomarkers = len(data_we_have.biomarker.unique())
if not os.path.exists(data_file):
print(f"Data file not found: {data_file}")
sys.exit(1) # Exit early if the file doesn't exist
else:
print(f"Data file found: {data_file}")
# Define the temporary result file
temp_result_file = os.path.join(temp_results_dir, f"temp_results_{j}_{r}_{m}.json")
dic = {}
if combstr not in dic:
dic[combstr] = []
accepted_order_dicts = metropolis_hastings_soft_kmeans(
data_we_have,
iterations,
n_shuffle,
)
utils.save_heatmap(
accepted_order_dicts,
burn_in,
thining,
folder_name=heatmap_folder,
file_name=f"{img_filename}",
title=f'heatmap of {filename}')
most_likely_order_dic = utils.obtain_most_likely_order_dic(
accepted_order_dicts, burn_in, thining)
most_likely_order = list(most_likely_order_dic.values())
tau, p_value = kendalltau(most_likely_order, range(1, n_biomarkers + 1))
dic[combstr].append(tau)
# Write the results to a unique temporary file inside the temp folder
with open(temp_result_file, "w") as file:
json.dump(dic, file, indent=4)
print(f"{filename} is done! Results written to {temp_result_file}")Current working directory: /Users/hongtaoh/Desktop/github/ebmBook
Data directory: /Users/hongtaoh/Desktop/github/ebmBook/data
Temp results directory: /Users/hongtaoh/Desktop/github/ebmBook/soft_kmeans/temp_json_results
Image directory: /Users/hongtaoh/Desktop/github/ebmBook/soft_kmeans/img
Processing with j=200, r=0.75, m=3
Data file found: /Users/hongtaoh/Desktop/github/ebmBook/data/150|200_3.csv/var/folders/wx/xz5y_06d15q5pgl_mhv76c8r0000gn/T/ipykernel_12345/1007084789.py:261: RuntimeWarning: overflow encountered in exp
prob_of_accepting_new_order = np.exp(iteration 10 done, current accepted likelihood: -4558.471198243365, current acceptance ratio is 100.00 %, current accepted order is dict_values([5, 9, 3, 4, 2, 1, 6, 8, 7, 10]),
150|200_3 is done! Results written to /Users/hongtaoh/Desktop/github/ebmBook/soft_kmeans/temp_json_results/temp_results_200_0.75_3.json8.2 Result
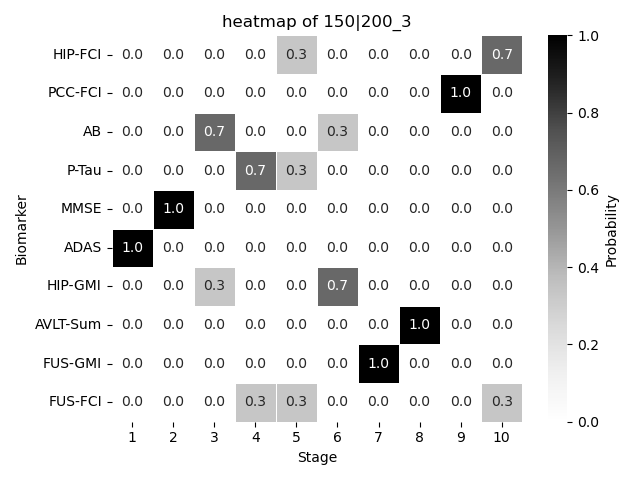
We plot the resulting \(S\) probalistically using a heatmap.
dic{'150|200': [-0.15555555555555553]}